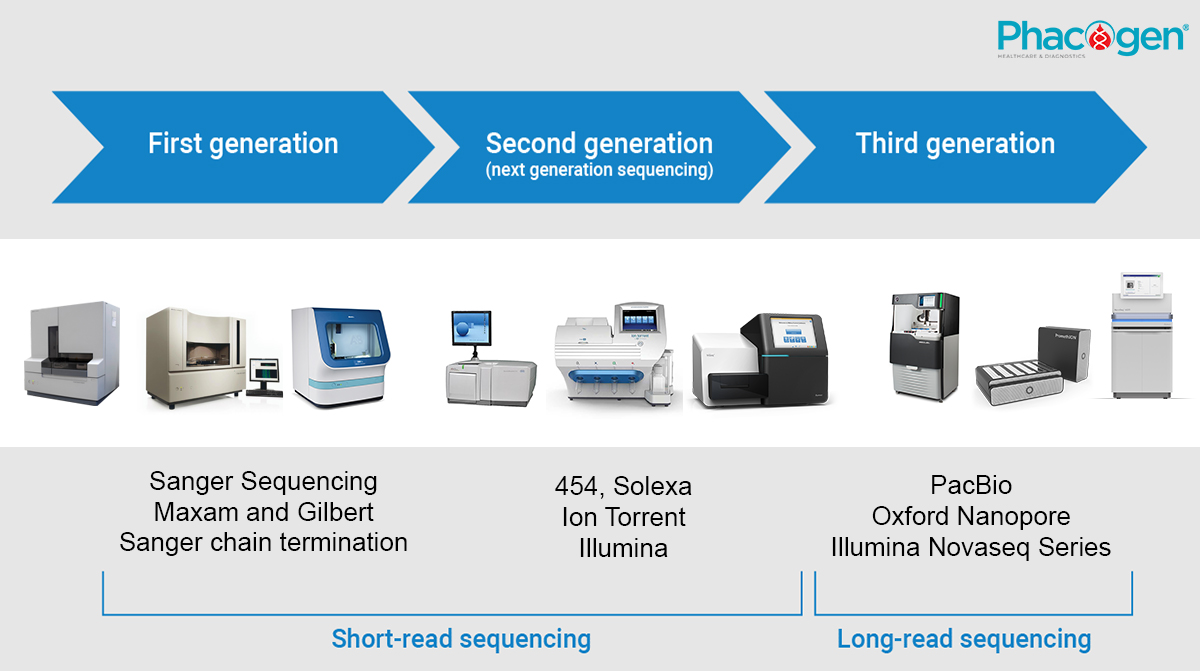
1. The first-generation gene sequencing - marking the beginning of the genomic era.
The Sanger sequencing method is the first-generation DNA sequencing method, developed by Frederick Sanger and colleagues in 1977. In the 1980s, Sanger's original method was automated by scientists at Caltech and commercialized by Applied Biosystems.
The method involves four test tubes, each containing a different labeled ddNTP (dideoxynucleotide triphosphate), along with regular dNTPs (deoxynucleotide triphosphates), for synthesis reactions. The products from the four tubes are then subjected to electrophoresis on a gel to observe DNA bands. The sequence of nucleotides is determined by the positions of the observed bands on the gel, read from the bottom to the top in the 5'-3' direction.
Today, automated sequencers are used to read the DNA sequence bands. The automated DNA sequencing technique, known as Dye Terminator Sequencing, utilizes fluorescently labeled ddNTPs to rapidly and efficiently sequence DNA. Each ddNTP is labeled with a different fluorescent dye, allowing sequencing to be performed in a single reaction. Currently, automated Sanger sequencing technology is still used, primarily in clinical laboratories where lower throughput, higher per-sample cost, and longer read lengths of 500-1,000 bp are acceptable.
2. Second-generation sequencing, also known as short-read sequencing, became rapidly efficient.
After acquiring Solexa, Illumina made significant advancements in the development of Next-Generation Sequencing (NGS) technology. A key feature of Illumina's NGS platform is "bridge amplification," which allows for the generation of dense clusters of amplified fragments on a silicon chip. The sequencing is based on the principle of synthesis, known as Sequencing by Synthesis (SBS), where the original DNA molecule is amplified into a large cluster of copies, enabling the detection of fluorescent signals upon the addition of individual dNTPs.
Over time, the number of clusters that can be read has significantly increased, making Illumina's devices successful in commercializing high-throughput parallel sequencing technology. NGS is currently the predominant sequencing technology used. Its high capacity allows for cost-effective sequencing. However, NGS has limitations in read length; NGS platforms typically generate reads of around 50-500 bp, which is suitable for resequencing projects, SNP calling, and sequencing of short target amplicons.
3. Third-generation sequencing - the rise of long-read sequencing
Short-read sequencing is not suitable for all genome sequencing projects. The single-molecule real-time sequencing (SMRT) technique developed by PacBio is performed in a nano-scale pore with unique properties that allow the observation of single-molecule fluorescence signals as fluorescently labeled nucleotides are incorporated into the growing DNA strand in the nano-scale zero-mode waveguide.
Another method for generating long reads is based on nanopore sequencing technology developed by Oxford Nanopore Technologies (ONT). The latest versions of their single-molecule sequencing systems have been significantly miniaturized. Systems such as GridION, MinION, or Flongle from Oxford Nanopore Technologies are handheld devices capable of sequencing RNA and DNA with data outputs of over 2 Mb.
Illumina has also introduced the Novaseq series, which represents a new peak in sequencing power for commercial platforms. The NovaSeq 6000 (S4 flow cell) can generate outputs of up to 3000 Gb per run (Illumina, 2017). Illumina's long-read sequencing devices are being developed for the future, promising increased flexibility and scalability, thereby reducing the cost of whole-genome sequencing.
Reference Source: