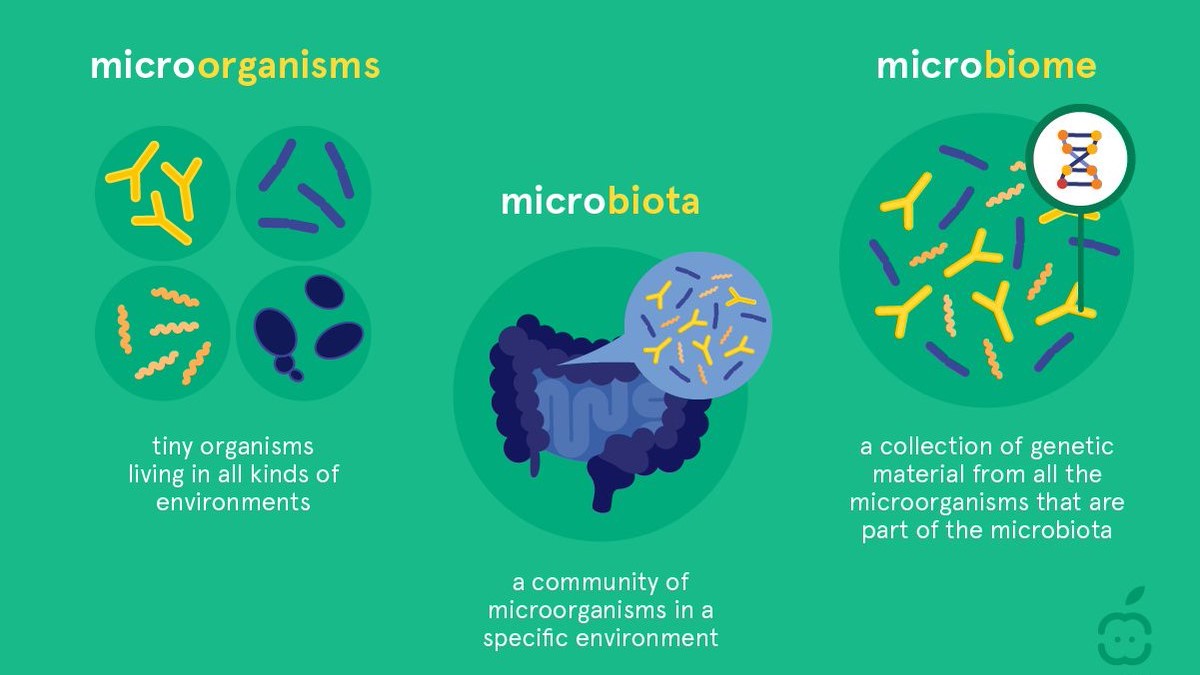
A microbiota is a symbiotic, symbiotic microflora living in soil, atmosphere, oceans, plants and animals, or in humans. Microorganisms can be bacteria, viruses, fungi or protozoa, which interact with the host organism or the ecosystem in which they reside, producing positive/negative/no affection for the host as well as other organisms and their habitats.
For example, the composition of the human gut microbiota is relevant to physiological and psychological health, and the soil microbiome can influence crop yield.
Microbiome is a term that describes the genomes of microorganisms in the microbiota. In humans, there are about 300 million genes in the microbiome, 150 times more than the total number of human genes. Up to date, approximately 3.3 million genes encoding proteins have been sequenced.
 Metagenomics is the study of Microbiome (genome of microorganisms) by using PCR technique combined with different sequencing technologies and bioinformatics technology to analyze the microbial genome in any any natural environment.
Metagenomics is the study of Microbiome (genome of microorganisms) by using PCR technique combined with different sequencing technologies and bioinformatics technology to analyze the microbial genome in any any natural environment.
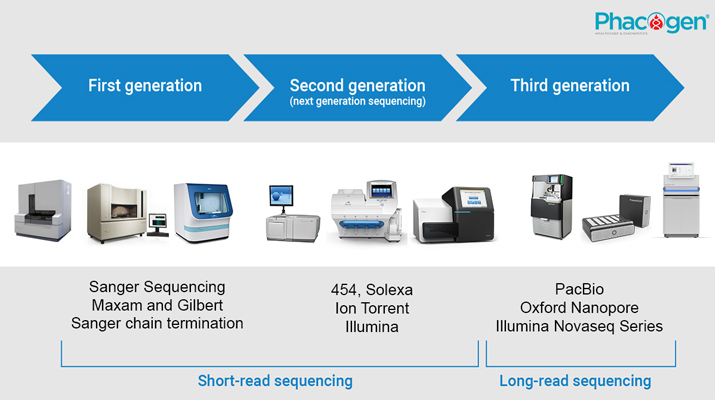
Traditional microbiological methods do not allow the presence of non-culturable microorganisms such as anaerobes. Furthermore, conventional microbiology also does not allow us to determine the abundance of bacteria in the biota. Today, using next-generation sequencing (NGS) technology, scientists have overcome these limitations and better understand the role of each microbial species in an ecosystem.
Using the 16S-rRNA gene sequencing method by NGS technology helps people deeply understand the characteristics and roles of the microbiome of many habitats such as soil, water, plants and animals, especially bacteria in different parts of the body. The human microbiome accompanies us throughout life. Because of the important symbiotic relationship between us and our microbial communities, microorganisms are of great importance to health. For example, the human gut microbiota is considered an important organ of the human body, it has been shown to be directly related to a number of diseases such as allergies, and some metabolic diseases. All the genomic information in this microbiome is said to be the "second genome" of the human being.
Although the understanding of the microbiome in the habitats in the developed countries is increasingly clear and has many applications in health care, plants and animals, the scientific research about 'microbiota' in our country has not really been noticed. Because the "microbiota" depends on many factors such as host, geographical area, food, water source, the "microbiota" of any individual or habitat has its own endemicity. The construction, development and application of knowledge about the microbiota of humans, animals and the environment for application in health care, navigating beneficial habitats for humans, livestock and plants will be an industry. Science promises to explode in the next few years.
Phacogen Institute of Technology uses Illumina's next-generation sequencing technology (NGS) and metagenomic approach (analysis of all general genomic information in the microbiome) to help researchers overcome the limitations of tedious traditional methods for isolating and culturing single species of microorganisms. The measurement approach can also help us effectively analyze the entire microbial community and provide insights into the relationship between humans and microorganisms and new ideas on many biomedical issues.
Those who are interested in Metagenomics should take a look the course "Application of NGS-Illumina in 16s Metagenomics Analysis" - The FIRST and ONLY Metagenomics Course IN VIETNAM up to now.
For more information, please CLICK HERE